Decoding the Daily Metabolic Cycle with Quantitative Proteomics: Insights from a Tiny Alga
Dr Sougata Roy, Assistant Professor of Biology at Ashoka University, and his research group investigate how the cell-based internal clock regulates metabolism, using advanced techniques like quantitative proteomics. This work offers important insights into how cells stay in tune with the day-night cycle and could have broader implications across species.
Almost every organism living on planet Earth experiences the daily oscillations of light and temperature throughout the day. To thrive in this dynamic environment, organisms must use resources optimally by performing key metabolic functions at specific times of the day. This generates a daily rhythmic pattern in metabolic activities, which is governed by a cell-based, self-sustaining biochemical oscillator known as the circadian clock. A key function of the circadian clock is to anticipate the daily environmental changes, such as light-dark transitions, and help organisms synchronise their activities accordingly. Such cellular clocks are widespread, from bacteria to humans. One of the most well-known examples of clock-controlled activity in humans is the sleep–wake cycle.
Understanding the molecular mechanisms by which the circadian clock regulates metabolism is a fundamental question that has garnered widespread interest across diverse species. This was the primary motivation for Dr Sougata Roy and his research group to explore the lesser-studied area using high-throughput circadian proteomics. Proteins are synthesised from their parent biomolecule, the mRNA (Messenger RNA). Enzymes, a class of proteins, are essential catalysts in metabolic pathways. Until recently, daily metabolic rhythms were primarily attributed to the clock’s ability to regulate mRNAs. It is well known that clock regulation of mRNAs often does not translate to protein expression, as proteins can be directly regulated by the clock, bypassing their parent mRNAs. Dr Roy’s team hypothesised that daily regulation of metabolic enzymes can drive the overall rhythmicity of cellular metabolism. They envisaged that capturing this proteome dynamics would be a robust approach to understanding how metabolic functions change over the daily cycle. Hence, the research aimed not only to uncover the molecular mechanisms underlying well-established metabolic rhythms but also to identify previously unrecognised metabolic rhythms occurring over the daily cycle.
To accomplish this, Dr Roy’s group studied Chlamydomonas reinhardtii. Chlamydomonas reinhardtii, a unicellular, photosynthetic, and motile member of the phytoplankton clade, maintains a precise circadian program. During the day, it produces the apparatus to capture and convert sunlight to chemical energy, fuelling its cellular processes. However, UV-sensitive processes, such as DNA synthesis and cell division, are programmed at night. Although it is a unicellular system, some of its primary metabolic pathways are analogous to multicellular organisms, making it an ideal model to reveal how the circadian dynamics of the proteome influences the metabolic rhythms across multiple species.
The hallmark of circadian clock-regulated processes is that they continue to be rhythmic even under constant conditions (constant light or darkness). Dr Roy’s work involved culturing cells under normal day-night cycles to synchronise their internal clocks, followed by transferring them to constant conditions to capture circadian clock-driven changes. They collected the cells at regular intervals continuously for two days, later processing and quantifying the harvested proteins from these cells using high-throughput mass-spectrometry-based proteomics, an approach which helps to identify proteins and quantify their abundance in the cell.
Their analysis revealed the temporal pattern of protein abundance across the day-night cycle to be widespread. Approximately half of the quantified proteins peaked during the day, and about one-third peaked at night. Functional categorisation of these proteins revealed that enzymes involved in amino acid (AA) biosynthesis, tricarboxylic acid cycle, and fatty-acid (FA) biosynthesis were predominantly peaking during the night. On the other hand, enzymes and proteins associated with photosynthesis, light sensitivity, and redox metabolism were more abundant during the day (Figure 1). This coordinated temporal accumulation of proteins indicated that many novel metabolic processes (e.g. BOX C/D RNA modification system, AA metabolism, and FA metabolism) are driven by the circadian clock.
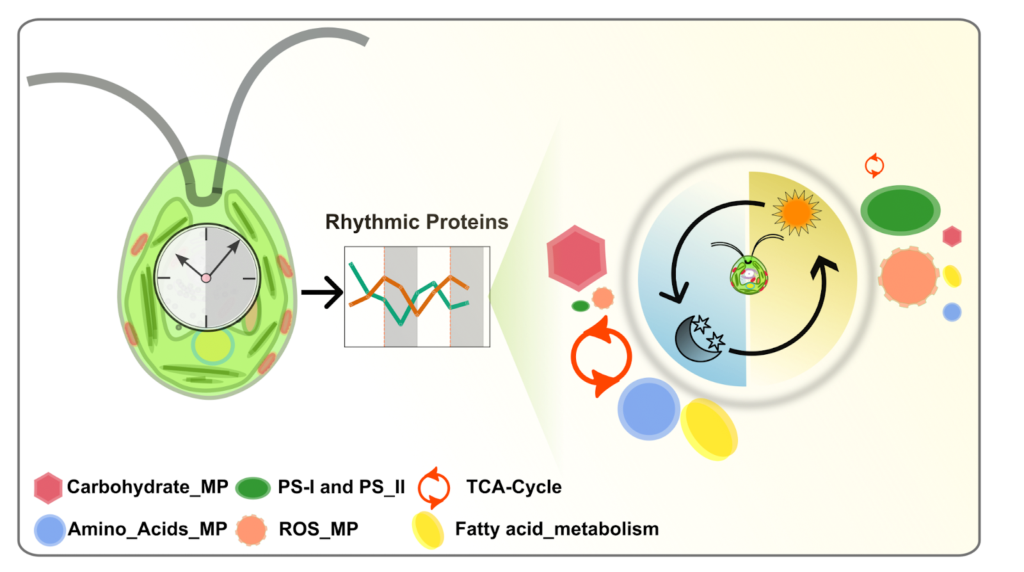
These findings suggest that clock regulation of the proteome is widespread, and it plays a crucial role in driving the daily rhythms of key metabolic pathways. Chlamydomonas is a microscopic plant with many animal-like characteristics, which makes it a valuable model for understanding the circadian regulation of metabolism across species. Future research could focus on a deeper investigation of the individual target enzymes and proteins identified in this study to elucidate their specific roles in the daily regulation of metabolism. Additionally, comparative analyses across diverse species may uncover conserved and divergent features of circadian metabolic regulation, providing insights that could have applications in agriculture, bioenergy, and human health.
– Edited by Kangna Verma, Academic Communications, Research and Development Office
Reference Article:
Circadian Proteomics Reassesses the Temporal Regulation of Metabolic Rhythms by Chlamydomonas Clock
https://doi.org/10.1111/pce.15354
Authors : Dinesh Balasaheb Jadhav, Sougata Roy
Study at Ashoka